...
| Panel | ||
|---|---|---|
| ||
Q. What is different about SST between the two ensemble forecasts? | ||
| Panel | title |
Cross-sections of a ensemble
...
member
To show a cross-section of a particular ensemble member, use the macro
...
ens_xs.mv
...
.
This works in the same way as the
...
hres_xs.mv macros.
...
...
Identifying sensitive
...
regions
Find ensemble members that appear to produce a better forecast and look to see how the initial development in these members differs.
- Select 'better' forecasts using the stamp plots and use ens_to_an.mv to modify the list of ensemble plots.
- Use pf_to_cf_diff and ens_to_an_diff to take the difference between these perturbed ensemble member forecasts from the control and analyses to also look at this.
| Panel | ||
|---|---|---|
| ||
Q. Can you tell which area is more sensitive for the forecast? |
...
Task
...
4: Cumulative distribution function, probabilities and percentiles
Recap
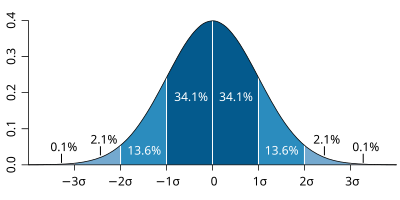
| The probability distribution function of the normal distribution or Gaussian distribution. The probabilities expressed as a percentage for various widths of standard deviations (σ) represent the area under the curve. |
|---|
Figure from Wikipedia. |
...
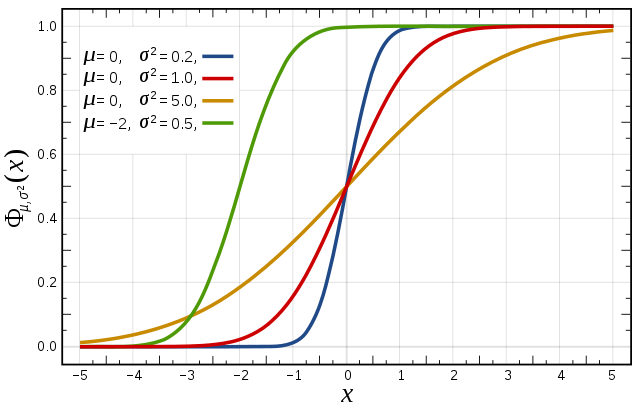
Cumulative distribution function for a normal |
|---|
Figure from Wikipedia. |
Cumulative distribution function (CDF)
The figures above illustrate the relationship between a normal distribution and its associated cumulative distribution function. The CDF is constructed from the area under the probability density function.
...
| Panel | ||
|---|---|---|
| ||
Q. Compare the CDF from the different forecast ensembles; what can you say about the spread? |
Exercise
...
The paper by Pantillon et al, describes the use of clustering to identify the main scenarios among the ensemble members.
This exercise repeats some of the plots from the previous one but this time with clustering enabled.
Using clustering will highlight the ensemble members in each cluster in the plots.
In this exercise you will:
- Construct your own qualitative clusters by choosing members for two clusters
- Generate clusters using principal component analysis (similar to Pantillon et al).
Task 1: Create your own clusters
Clusters can be created manually from lists of the ensemble members.
Choose members for two clusters. The stamp maps are useful for this task.
From the stamp map of z500 at 24/9/2012 (t+96), identify ensemble members that represent the two most likely forecast scenarios.
It is usual to create clusters from z500 as it represents the large-scale flow and is not a noisy field. However, for this particular case study, the stamp map of 'tp' (total precipitation) over France is also very indicative of the distinct forecast scenarios.
| Panel | ||
|---|---|---|
| ||
Right-click 'ens_oper_cluster.example.txt' and select Edit (or make a duplicate) The file contains two example lines:
The first line defines the list of members for 'Cluster 1': in this example, members 2, 3, 4, 9, 22, 33, 40. The second line defines the list of members for 'Cluster 2': in this example, members 10, 11, 12, 31, 49. Change these two lines!. You can create multiple cluster definitions by using the 'Duplicate' menu option to make copies of the file for use in the plotting macros.. The filename is important! |
| Panel | ||
|---|---|---|
| ||
Use the clusters of ensemble members you have created in Set Replot ensembles:RMSE: plot the RMSE curves using Stamp maps: the stamp maps will be reordered such at the ensemble members will be groups according to their cluster. Applies to Spaghetti maps: with clusters enabled, two additional maps are produced which show the contour lines for each cluster. The spaghetti maps are similar to Figure 10. in Pantillon et al. |
| Panel | |||||
|---|---|---|---|---|---|
| |||||
The macro Use If your cluster definition file is called 'ens_oper_cluster.example.txt', then Edit
If your cluster definition file is has another name, e.g. ens_oper_cluster.fred.txt, then members_1=["cl.fred.1"]. Plot other parameters:Plot total precipitation for France ( |
| Panel | ||
|---|---|---|
| ||
Q. Experiment with the choice of members in each clusters and plot z500 at t+96 (Figure 7 in Pantillon et al.). How similar are your cluster maps? |
Task 2: Empirical orthogonal functions / Principal component analysis
A quantitative way of clustering an ensemble is by computing empirical orthogonal functions from the differences between the ensemble members and the control forecast.
Although geopotential height at 500hPa at 00 24/9/2012 is used in the paper by Pantillon et al., the steps described below can be used for any parameter at any step.
The eof.mv macro computes the EOFs and the clustering.
| Warning |
|---|
Always use the Otherwise cluster_to_an.mv and other plots with clustering enabled will fail or plot with the wrong clustering of ensemble members. If you change step or ensemble, recompute the EOFS and cluster definitions using eof.mv. Note however, that once a cluster has been computed, it can be used for all steps with any parameter. |
| Panel | ||
|---|---|---|
| ||
Edit 'eof.mv' Set the parameter to use, choice of ensemble and forecast step required for the EOF computation:
Run the macro. The above example will compute the EOFs of geopotential height anomaly at 500hPa using the 2012 operational ensemble at forecast step 00Z on 24/09/2012. A plot will appear showing the first two EOFs (similar to Figure 5 in Pantillon et al.) The geographical area for the EOF computation is: 35-55N, 10W-20E (same as in Pantillon et al). If desired it can be changed in |
| Panel | |||||||
|---|---|---|---|---|---|---|---|
| |||||||
The eof.mv macro will create a text file with the cluster definitions, in the same format as described above in the previous task. The filename will be different, it will have 'eof' in the filename to indicate it was created by using empirical orthogonal functions.
If a different ensemble forecast is used, for example This cluster definition file can then be used to plot any variable at all steps (as for task 1). |
| Panel | ||
|---|---|---|
| ||
Q. What do the EOFs plotted by eof.mv show? |
| Panel | |||||
|---|---|---|---|---|---|
| |||||
Use the cluster definition file computed by eof.mv to the plot ensembles and maps with clusters enabled (as described for task 1, but this time with the 'eof' cluster file). The macro Use Edit
Run the macro. If time also look at the total precipitation (tp) over France and PV/320K. |
5. Percentiles and probabilities
To further compare the 2012 and 2016 ensemble forecasts, plots showing the percentile amount and probabilities above a threshold can be made for total precipitation.
Use these icons:
Both these macros will use the 6-hourly total precipitation for forecast steps at 90, 96 and 102 hours, plotted over France.
Task 1. Plot percentiles of total precipitation
Edit the percentile_tp_compare.mv icon.
Set the percentile for the total precipitation to 75%:
| Code Block | ||
|---|---|---|
| ||
#The percentile of ENS precipitation forecast
perc=75 |
Run the macro and compare the percentiles from both the forecasts. Change the percentiles to see how the forecasts differ.
Task 2: Plot probabilities of total precipitation
This macro will produce maps showing the probability of 6-hourly precipitation for the same area as in Task 1.
In this case, the maps show the probability that total precipitation exceeds a threshold expressed in mm.
Edit the prob_tp_compare.mv and set the probability to 20mm:
| Code Block | ||
|---|---|---|
| ||
#The probability of precipitation greater than
prob=20 |
Run the macro and view the map. Try changing the threshold value and run.
| Panel | ||
|---|---|---|
| ||
Q. Using these two macros, compare the 2012 and 2016 forecast ensemble. Which was the better forecast for HyMEX flight planning? |
Exercise 4: Cluster analysis
The paper by Pantillon et al, describes the use of clustering to identify the main scenarios among the ensemble members.
This exercise repeats some of the plots from the previous one but this time with clustering enabled.
Using clustering will highlight the ensemble members in each cluster in the plots.
In this exercise you will:
- Construct your own qualitative clusters by choosing members for two clusters
- Generate clusters using principal component analysis (similar to Pantillon et al).
Task 1: Create your own clusters
Clusters can be created manually from lists of the ensemble members.
Choose members for two clusters. The stamp maps are useful for this task.
From the stamp map of z500 at 24/9/2012 (t+96), identify ensemble members that represent the two most likely forecast scenarios.
It is usual to create clusters from z500 as it represents the large-scale flow and is not a noisy field. However, for this particular case study, the stamp map of 'tp' (total precipitation) over France is also very indicative of the distinct forecast scenarios.
| Panel | ||
|---|---|---|
| ||
Right-click 'ens_oper_cluster.example.txt' and select Edit (or make a duplicate) The file contains two example lines:
The first line defines the list of members for 'Cluster 1': in this example, members 2, 3, 4, 9, 22, 33, 40. The second line defines the list of members for 'Cluster 2': in this example, members 10, 11, 12, 31, 49. Change these two lines!. You can create multiple cluster definitions by using the 'Duplicate' menu option to make copies of the file for use in the plotting macros.. The filename is important! |
| Panel | ||
|---|---|---|
| ||
Use the clusters of ensemble members you have created in Set Replot ensembles:RMSE: plot the RMSE curves using Stamp maps: the stamp maps will be reordered such at the ensemble members will be groups according to their cluster. Applies to Spaghetti maps: with clusters enabled, two additional maps are produced which show the contour lines for each cluster. The spaghetti maps are similar to Figure 10. in Pantillon et al. |
| Panel | |||||
|---|---|---|---|---|---|
| |||||
The macro Use If your cluster definition file is called 'ens_oper_cluster.example.txt', then Edit
If your cluster definition file is has another name, e.g. ens_oper_cluster.fred.txt, then members_1=["cl.fred.1"]. Plot other parameters:Plot total precipitation for France ( |
| Panel | ||
|---|---|---|
| ||
Q. Experiment with the choice of members in each clusters and plot z500 at t+96 (Figure 7 in Pantillon et al.). How similar are your cluster maps? |
Task 2: Empirical orthogonal functions / Principal component analysis
A quantitative way of clustering an ensemble is by computing empirical orthogonal functions from the differences between the ensemble members and the control forecast.
Although geopotential height at 500hPa at 00 24/9/2012 is used in the paper by Pantillon et al., the steps described below can be used for any parameter at any step.
The eof.mv macro computes the EOFs and the clustering.
| Warning |
|---|
Always use the Otherwise cluster_to_an.mv and other plots with clustering enabled will fail or plot with the wrong clustering of ensemble members. If you change step or ensemble, recompute the EOFS and cluster definitions using eof.mv. Note however, that once a cluster has been computed, it can be used for all steps with any parameter. |
| Panel | ||
|---|---|---|
| ||
Edit 'eof.mv' Set the parameter to use, choice of ensemble and forecast step required for the EOF computation:
Run the macro. The above example will compute the EOFs of geopotential height anomaly at 500hPa using the 2012 operational ensemble at forecast step 00Z on 24/09/2012. A plot will appear showing the first two EOFs (similar to Figure 5 in Pantillon et al.) The geographical area for the EOF computation is: 35-55N, 10W-20E (same as in Pantillon et al). If desired it can be changed in |
| Panel | |||||||
|---|---|---|---|---|---|---|---|
| |||||||
The eof.mv macro will create a text file with the cluster definitions, in the same format as described above in the previous task. The filename will be different, it will have 'eof' in the filename to indicate it was created by using empirical orthogonal functions.
If a different ensemble forecast is used, for example This cluster definition file can then be used to plot any variable at all steps (as for task 1). |
| Panel | ||
|---|---|---|
| ||
Q. What do the EOFs plotted by eof.mv show? |
| Panel | |||||
|---|---|---|---|---|---|
| |||||
Use the cluster definition file computed by eof.mv to the plot ensembles and maps with clusters enabled (as described for task 1, but this time with the 'eof' cluster file). The macro Use Edit
Run the macro. If time also look at the total precipitation (tp) over France and PV/320K. |
| Panel | ||
|---|---|---|
| ||
Q. How similar is the PCA computed clusters to your manual clustering? |
| Panel | ||
|---|---|---|
| ||
For those interested: The code that computes the clusters can be found in the Python script: This uses the 'ward' cluster method from SciPy. Other cluster algorithms are available. See http://docs.scipy.org/doc/scipy/reference/generated/scipy.cluster.hierarchy.linkage.html#scipy.cluster.hierarchy.linkage The python code can be changed to a different algorithm or the more adventurous can write their own cluster algorithm! |
| Panel | ||
|---|---|---|
| ||
Q. How similar is the PCA computed clusters to your manual clustering? |
| Panel | ||
|---|---|---|
| ||
For those interested: The code that computes the clusters can be found in the Python script: This uses the 'ward' cluster method from SciPy. Other cluster algorithms are available. See http://docs.scipy.org/doc/scipy/reference/generated/scipy.cluster.hierarchy.linkage.html#scipy.cluster.hierarchy.linkage The python code can be changed to a different algorithm or the more adventurous can write their own cluster algorithm! |
Exercise 5. Percentiles and probabilities
To further compare the 2012 and 2016 ensemble forecasts, plots showing the percentile amount and probabilities above a threshold can be made for total precipitation.
Use these icons:
Both these macros will use the 6-hourly total precipitation for forecast steps at 90, 96 and 102 hours, plotted over France.
Task 1. Plot percentiles of total precipitation
Edit the percentile_tp_compare.mv icon.
Set the percentile for the total precipitation to 75%:
| Code Block | ||
|---|---|---|
| ||
#The percentile of ENS precipitation forecast
perc=75 |
Run the macro and compare the percentiles from both the forecasts. Change the percentiles to see how the forecasts differ.
Task 2: Plot probabilities of total precipitation
This macro will produce maps showing the probability of 6-hourly precipitation for the same area as in Task 1.
In this case, the maps show the probability that total precipitation exceeds a threshold expressed in mm.
Edit the prob_tp_compare.mv and set the probability to 20mm:
| Code Block | ||
|---|---|---|
| ||
#The probability of precipitation greater than
prob=20 |
Run the macro and view the map. Try changing the threshold value and run.
| Panel | ||
|---|---|---|
| ||
Q. Using these two macros, compare the 2012 and 2016 forecast ensemble. Which was the better forecast for HyMEX flight planning? |
Exercise 6. Assessment of forecast errors
...