Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
Terminology
The CPDN OpenIFS@Home documentation uses slightly different terminology to what OpenIFS users might be used to.
OpenIFS@Home Ancillary Files
This refers to all the input files that the OpenIFS model needs to run:
- initial data to start a forecast (normally obtained from ECMWF);
- ifsdata and climate files (downloaded from the OpenIFS ftp site);
- model namelist (only parts of the OpenIFS namelist can be uploaded, see below)
These ancillary files are uploaded in a zipped format to the CPDN repository.
Experiment information is read from the initial files GRIB metadata upon upload to the OpenIFS@Home repository and used to set the experiment id, date/time of forecast etc.
Experiment namelist
It's not possible to upload a complete model namelist to the OpenIFS@Home website. This is because some restrictions are necessary in order for the model to work correctly and efficiently when running on a volunteer's computer.
At the time of writing, the only part of the model namelist allowed to be uploaded is the 'NAMFPC', or 'FullPos' namelist, which controls all the model's output. Even this is vetted by CPDN first to ensure the model does not produce more output than can be reasonably handled by the volunteer's computer and network for transfer of the results back to the CPDN database.
Apart from the model's NAMFPC output control namelist, the rest of the namelist is selected as a 'template' when the experiment on OpenIFS@Home is configured. Namelist experiment templates exist for different resolutions and different classes of experiments; for example 10 day NWP forecasts, seasonal forecasts etc. For more details please see the OpenIFS@Home dashboard and documentation, or contact the CPDN team directly.
Preparing Initial Files to Upload
The upload of initial files is made from the OpenIFS@Home OpenIFS@home Dashboard on the OpenIFS@Home portal. It expects the initial files to be laid out in a particular directory structure.
There are 3 types of permitted directory structures as illustrated in the diagram left. They correspond to whether multiple dates are present, and whether ensembles of initial data are being used.
File format
Following the directory layout below, create a gzipped tarfile of the data ready to be uploaded.
| Code Block | ||||
|---|---|---|---|---|
| ||||
% tar cvfz hijc_2017041212.tgz hijc |
(a) Single date.
In this case the directory structure would be as following:
| Code Block |
|---|
% ls -R hijc # "hha4" is the experiment id in this example hijc/2017041212: ICMCLhijcINIT ICMSHhijcINIT specwavein wam_subgrid_0 ICMGGhijcINIT cdwavein uwavein wam_subgrid_1 ICMGGhijcINIUA sfcwindin wam_grid_tables wam_subgrid_2 |
| Info |
|---|
There is no fort.4 or namelist file and no 'ecmwf' directory as is common in the files downloaded from the ECMWF provided initial model files. |
(b) Multiple dates.
This case is an extension of (a) but with multiple sub-directories, one for each date. The example below shows two dates but it's possible to use many more.
| Code Block | ||
|---|---|---|
| ||
% ls -R hijc # "hha4" is the experiment id in this example hijc/2017041212: ICMCLhijcINIT ICMSHhijcINIT specwavein wam_subgrid_0 ICMGGhijcINIT cdwavein uwavein wam_subgrid_1 ICMGGhijcINIUA sfcwindin wam_grid_tables wam_subgrid_2 hijc/2017041300: ICMCLhijcINIT ICMSHhijcINIT specwavein wam_subgrid_0 ICMGGhijcINIT cdwavein uwavein wam_subgrid_1 ICMGGhijcINIUA sfcwindin wam_grid_tables wam_subgrid_2 |
(c) Ensemble of starts and multiple dates
This is the more complex case. Not only are there multiple dates but for each date there are multiple ensemble members.
The directory structure will now look like: <expid>/<date>/<ens>/.
The following examples shows two dates, each with 3 ensemble members. By convention, there is always a '000' member.
| Code Block | ||
|---|---|---|
| ||
% ls -R h76y # 'h76y' is the experiment id in this case h76y: 2016092500 h76y/2016092500: 000 001 002 h76y/2016092500/000: ICMCLh76yINIT ICMSHh76yINIT specwavein wam_subgrid_0 ICMGGh76yINIT cdwavein uwavein wam_subgrid_1 ICMGGh76yINIUA sfcwindin wam_grid_tables wam_subgrid_2 h76y/2016092500/001: ICMCLh76yINIT ...... # lines omitted for brevity h76y: 2016092512: h76y/2016092512: 000 001 002 h67y/2016092512/000: ICMCLh76yINIT ICMSHh76yINIT specwavein wam_subgrid_0 ..... # lines omitted for brevity |
| Info |
|---|
In this case, to save space, make links between common files in each ensemble directory. The files that are typically the same are the:
|
By making links we have a structure that looks like (omitting some files listed above for clarity):
| Code Block |
|---|
% ls -lR h76y ..... h76y/2016092500/000: total 32032 -rw-r----- 1 nagc rd 3055416 Jun 13 2019 ICMCLh76yINIT -rw-r----- 1 nagc rd 4489702 Jun 13 2019 ICMGGh76yINIT -rw-r----- 1 nagc rd 15648820 Jun 13 2019 ICMGGh76yINIUA -rw-r----- 1 nagc rd 9544080 Jun 13 2019 ICMSHh76yINIT -rw-r----- 1 nagc rd 2546565 Jun 13 2019 cdwavein -rw-r----- 1 nagc rd 6354333 Jun 13 2019 sfcwindin -rw-r----- 1 nagc rd 493848 Jun 13 2019 specwavein -rw-r----- 1 nagc rd 1138804 Jun 13 2019 uwavein -rw-r----- 1 nagc rd 5384833 Jun 13 2019 wam_grid_tables -rw-r----- 1 nagc rd 438485 Jun 13 2019 wam_subgrid_0 -rw-r----- 1 nagc rd 438485 Jun 13 2019 wam_subgrid_1 -rw-r----- 1 nagc rd 438485 Jun 13 2019 wam_subgrid_2 h76y/2016092500/001: total 29144 lrwxrwxrwx 1 nagc rd 20 Jun 13 2019 ICMCLh76yINIT -> ../000/ICMCLh76yINIT -rw-r----- 1 nagc rd 4487766 Jun 13 2019 ICMGGh76yINIT -rw-r----- 1 nagc rd 15755898 Jun 13 2019 ICMGGh76yINIUA -rw-r----- 1 nagc rd 9543716 Jun 13 2019 ICMSHh76yINIT -rw-r----- 1 nagc rd 2546565 Jun 13 2019 cdwavein -rw-r----- 1 nagc rd 6354333 Jun 13 2019 sfcwindin -rw-r----- 1 nagc rd 493848 Jun 13 2019 specwavein -rw-r----- 1 nagc rd 1138804 Jun 13 2019 uwavein lrwxrwxrwx 1 nagc rd 5384833 Jun 13 2019 wam_grid_tables -> ../000/wam_grid_tables lrwxrwxrwx 1 nagc rd 438485 Jun 13 2019 wam_subgrid_0 -> ../000/wam_subgrid_0 lrwxrwxrwx 1 nagc rd 438485 Jun 13 2019 wam_subgrid_1 -> ../000/wam_subgrid_1 lrwxrwxrwx 1 nagc rd 438485 Jun 13 2019 wam_subgrid_2 -> ../000/wam_subgrid_2 |
It can helpful to write a bash script to create these directory structures, check filesizes and links.
Search for available climate and ifsdata files
Normally all climate and ifsdata files will have been uploaded to CPDN for the supported resolutions.
To search what's available, go to the OpenIFS@Home Dashboard at: https://dev.cpdn.org/oifs_dashboard.php (login if required)
Click on the 'search' button:  .
.
ifsdata files
In the Type dropdown menu select 'ifsdata' to see the list of files available: 
This will show a list similar to:

To find out more information and download the file if required, click on the blue link.
climate files
In the Type dropdown menu select 'climate_data' to generate a similar list:

This will show a similar list from which individual files can be selected for more information and downloaded if required.

Preparing climate and ifsdata to upload
Normally, this will not be necessary. All the climate and ifsdata needed to run OpenIFS on the OpenIFS@Home platform will have already been uploaded.
New files would be needed if you plan to use a modified or different set of boundary forcings e.g. a modified sea-surface temperature.
In this case, it's advised that you contact the CPDN support team for help and advice in preparing these files.
| Panel | ||||||||
|---|---|---|---|---|---|---|---|---|
| ||||||||
|
| Panel | ||||||||
|---|---|---|---|---|---|---|---|---|
| ||||||||
|
Image Removed
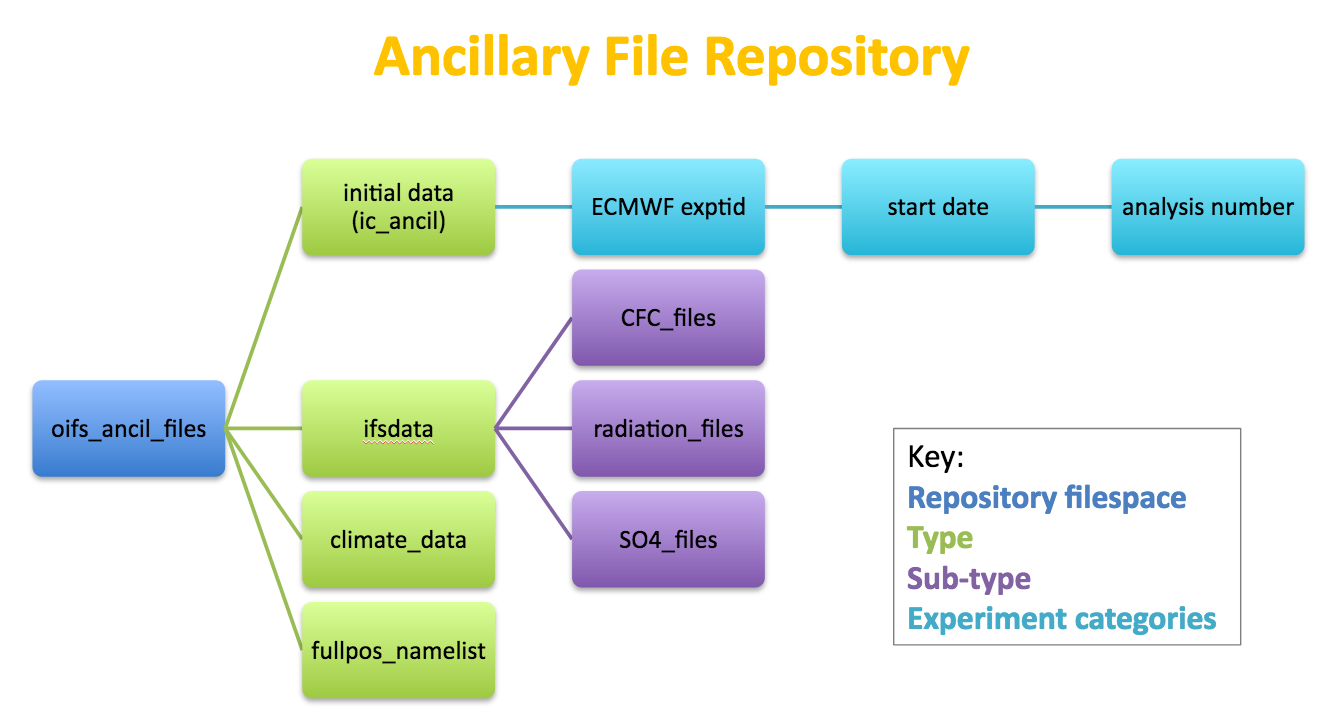 Image Added
Image AddedImage Removed
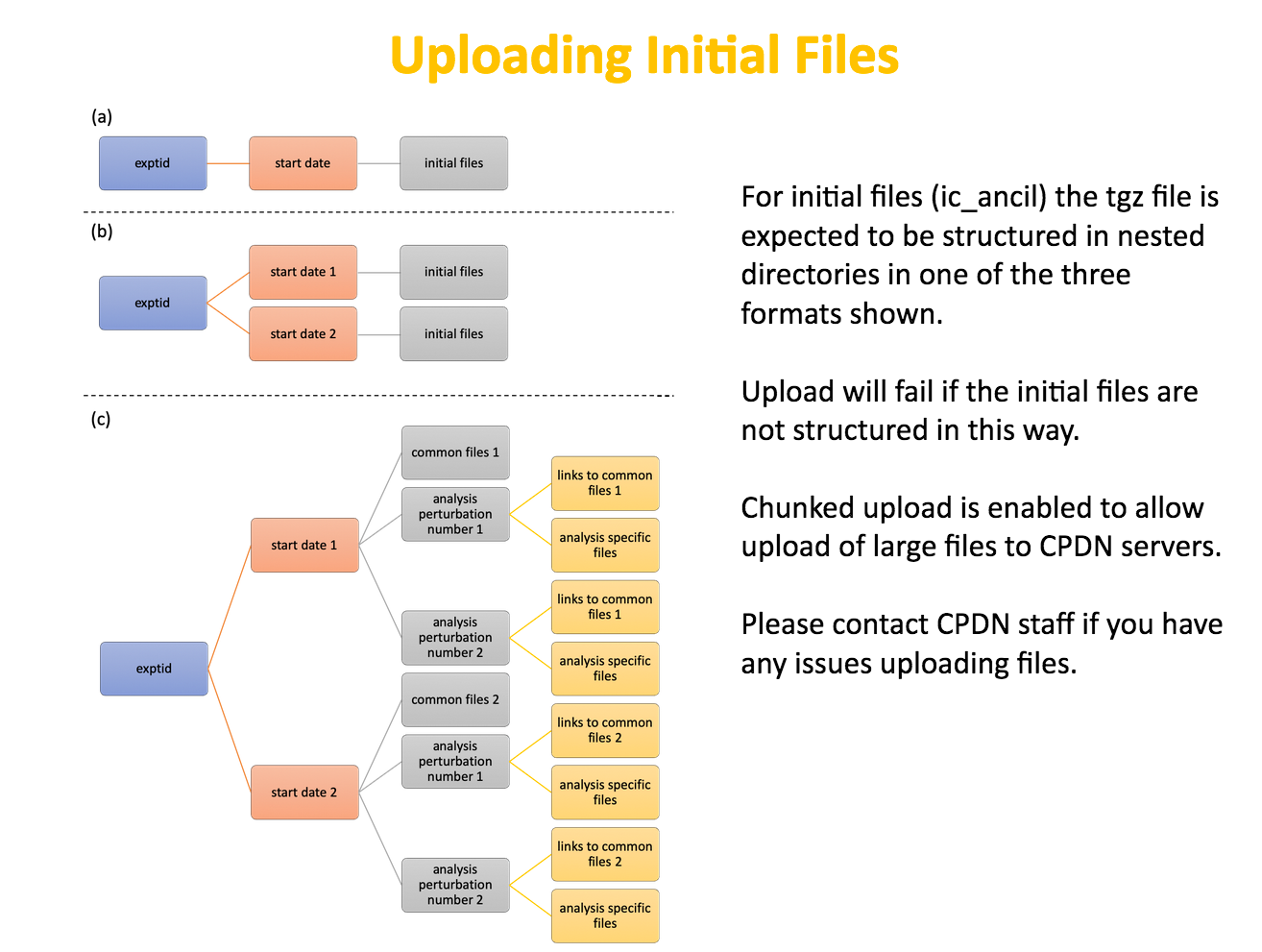 Image Added
Image Added