Opening a Jupyter Lab instanceYou can use ecinteractive to open up a Jupyter Lab instance on the HPCF. The application would effectively run on the allocated node for the job, and would allow you to conveniently interact with it from your browser. When running from VDI or your end user device, ecinteractive will try to open it in a new tab automatically. Alternatively you may manually open the URL provided to connect to your Jupyter Lab session. | No Format |
|---|
[user@aa6-100 ~]$ ecinteractive -j
Using interactive job:
CLUSTER JOBID STATE EXEC_HOST TIME_LIMIT TIME_LEFT MAX_CPUS MIN_MEMORY TRES_PER_NODE
aa 10225277 RUNNING aa6-104 6:00:00 5:58:07 2 8G ssdtmp:3G
To cancel the job:
/usr/local/bin/ecinteractive -k
Attaching to Jupyterlab session...
To manually re-attach go to http://aa6-104.ecmwf.int:33698/?token=b1624da17308654986b1fd66ef82b9274401ea8982f3b747 |
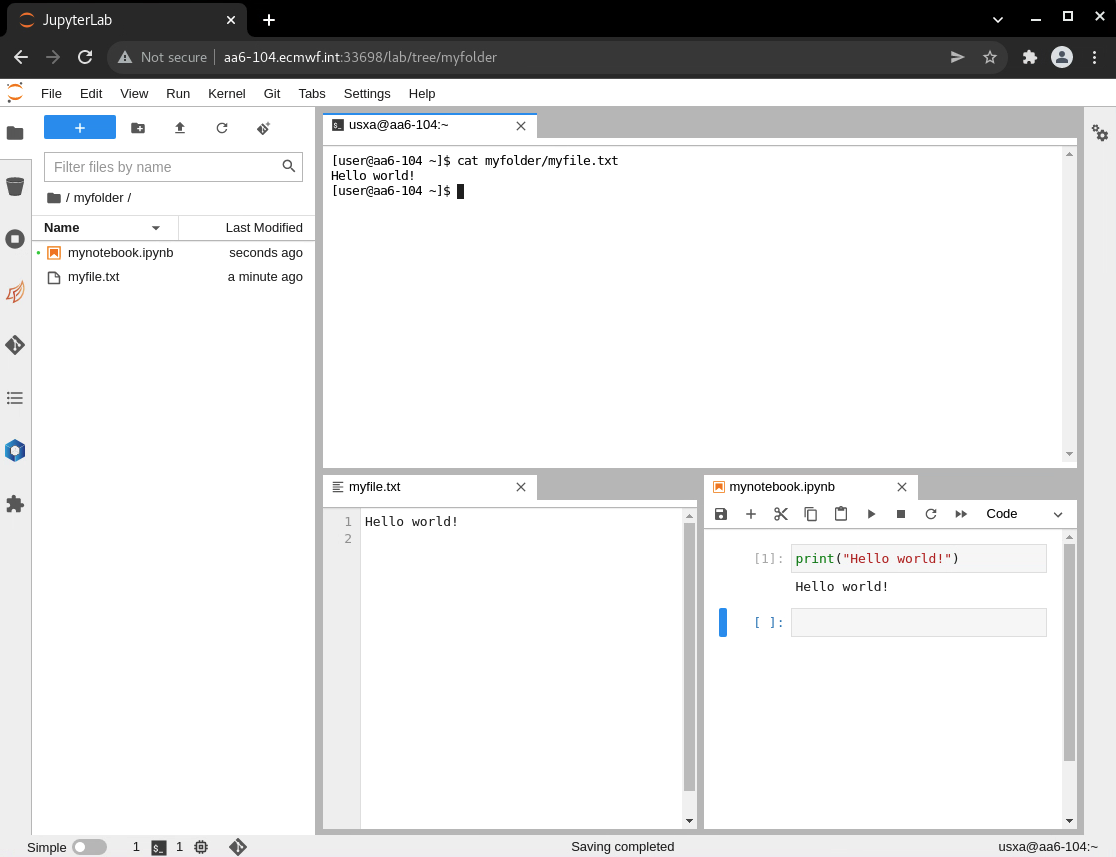 Image Modified Image Modified
To use your own conda environment as a kernel for Jupyter notebook you will need to have ipykernel installed in the conda environment before starting ecinteractive job. ipykernel can be installed with: | No Format |
|---|
[user@aa6-100 ~]$ conda activate {myEnv}
[user@aa6-100 ~]$ conda install ipykernel
[user@aa6-100 ~]$ python3 -m ipykernel install --user --name={myEnv} |
The same is true if you want to make your own Python virtual environment visible in Jupyterlab | No Format |
|---|
[user@aa6-100 ~]$ source {myEnv}/bin/activate
[user@aa6-100 ~]$ pip3 install ipykernel
[user@aa6-100 ~]$ python3 -m ipykernel install --user --name={myEnv} |
To remove your personal kernels from Jupyterlab once you don't need them anymore, you could do so with: | No Format |
|---|
jupyter kernelspec uninstall {myEnv} |
| Expand |
|---|
| title | How to connect to the Jupiter Lab instance from web browser on your end user device |
|---|
| - Authenticate with Teleport using jump.ecmwf.int as described in the Teleport SSH Access page
Copy ecinteractive script from hpc to your local machine:
| Code Block |
|---|
| scp username@hpc-login:/usr/local/bin/ecinteractive . |
Run ecinteractive script on your local machine:
| Code Block |
|---|
| ./ecinteractive -u username -j |
"-u" should be used to provide ECMWF username if username on your local machine doesn't match username on the Atos HPC. The ecinteractive job will start, new tab will automatically open on your web browser and attach to the Jupyter server running on the HPC. If not, you will get a link which can be used on your local machine only to paste it manually into the web browser. It should look like this: http://localhost:..... To kill your ecinteractive job from the local machine use:
| Code Block |
|---|
| ./ecinteractive -p hpc -k -u username |
|
HTTPS accessIf you wish to run Juptyer Lab on HTTPS instead of plain HTTP, you may use the -J option in ecinteractive. In that case, a personal SSL certificate would be created under ~/.ssl the first time, and would be used to encrypt the HTTP traffic between your browser and the compute node. In order to avoid browser security warnings, you may fetch the ~/.ssl/selfCA.crt certificate from the HPCF and import it into your browser as a trusted Certificate Authority. This is only needed once. | 
