For preparations and running the simulation needed for this tutorial click here ...
To start these tutorial please enter folder 'fwd'.
To plot a particular parameter and level we need to filter the desired fields from the resulting FLEXPART output GRIB file. Unfortunately, Metview's Grib Filter icon cannot handle these files (partly due to the local GRIB definition they use) so we need to use other means to cope with this task. For this reason and also to make FLEXPART output handling easier a set of Metview Macro Library Functions were developed. We will use these functions in the example below.
Inspecting the FLEXPART GRIB file
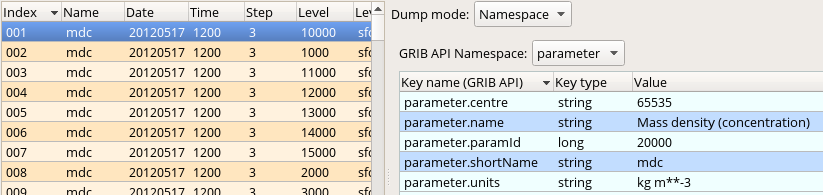
Before seeing the macro code to generate the plot we inspect the file itself we wanted to plot. Double-click on the 'conc_s001.grib' GRIB icon' in folder 'result_fw' to start up the Grib Examiner. We can see that this file contains the "mdc" (=Mass density concentration) fields we want to visualise. We can find out further details about this parameter by setting the Dump mode to Namespace and Namespace to Parameter:
Generating the plot
The macro to visualise the concentration fields on a given level is 'plot_conc.mv'.
In the macro first we define the level (8000 m) and the parameter ("mdc") we want to plot. Then we call the Macro Library Function mvl_flexpart_read_hl() to extract the data.
dIn="result_fwd_conc/" inFile=dIn & "conc_s001.grib" lev=8000 par="mdc" #Read fields on the given height level g=mvl_flexpart_read_hl(inFile,par,lev,-1,1)
Next, we define the contouring definition. The units we used here are ng m**-3 because for parameter "mdc" the native units (kg m**-3) are automatically scaled by the plotting library (see details about the this scaling for various FLEXPART GRIB fields here.
#The contour levels
cont_list=[1,10,50,100,150,200,250,500,750,1000,2000,6000]
#Define contour shading
conc_shade = mcont(
legend : "on",
contour : "off",
contour_level_selection_type : "level_list",
contour_level_list : cont_list,
contour_label : "off",
contour_shade : "on",
contour_shade_method : "area_fill",
contour_shade_max_level_colour : "red",
contour_shade_min_level_colour : "RGB(0.14,0.37,0.86)",
contour_shade_colour_direction : "clockwise",
contour_method : "linear"
)
Next, we build the title with mvl_flexpart_title(). Please note that we need to explicitly specify the plotting units!
title=mvl_flexpart_title(g,0.3,"ng m**-3")
Finally we define the view with the map:
and generate the plot:
plot(view,g,conc_shade,title)
Having run the macro we will get a plot like this (after navigating to step 39h):